Nano-particle Bio Conjugation
Relating to work conducted from 2008 to 2011
In the introduction to cell biology, non-specific endocytosis and receptor mediated endocytosis sections it has been discussed that the end goal of this project was to create nano-particles conjugated to proteins that can then be incubated with cells and used to track where the signalling proteins go and exactly what they do. Ultimately it would also be nice to produce nano-drug carriers, particles that can carry drugs to exactly the right places in the right cells so they can work without producing any unwanted side effects, but that is a much bigger undertaking.
In the non-specific endocytosis section an understanding was gained about how to stop nano-particles from entering cells when unwanted, and in the receptor mediated endocytosis section there was a discussion on how a genetically engineered protein called SST was tested and found to travel the same specific cellular pathways as the un-engineered protein, but to also have a second, fluorescent protein attached to it (RFP). As the newly created protein is genetically engineered it is a single protein that is both SST and RFP, and hence will not, for example, fall apart. It was also tested and proven to work efficiently to label SST receptors inside cells. The final test, therefore, was to attach a similar protein to a nano-particle. While this may sound easy, even a slight alteration to the protein structure can destroy the SST's activity. On top of this, we also have some other quite difficult specifications. As a nano-particle we wish to use a nano-diamond. This might sound extravagant but nano-sized pieces of diamond can be produced in large quantities but simply blowing up TNT (making them cheap), and tiny diamonds have many nice features. First, they are pure carbon, so are inert and non-toxic to cells, second, they can emit light through things called colour centres, and these are not damaged by the cells or bleached in time (as is the case for dyes). Finally, the carbon surfaces of the diamonds are a nice starting point for chemical bonding reactions. The second, more challenging, specification of the project was that we do not want to have to perfect a new conjugation procedure for every different protein we wish to attach to the diamond as this can be tricky and time consuming. Hence our aim was to optimise the process of attaching a single protein to the diamond. This protein by itself will not be very useful, but will have the property that it will attach incredibly strongly to a second protein in the same way that only the right key will fit in the right lock. Hence the aim was to produce lots of protein covered nano-diamonds, and then generically produce any protein of interest, pre-bound to the other half of the linker molecule. Finally, all we have to do is mix the two products together and we will end up with our required nano-particle covered in an active peptide of choice. This all sounds like a very complicated and long-winded way of doing things, however, it is, in fact, about the easiest way that ensures that all the components will work at the end. The image, right, shows a schematic of the scheme envisaged, here attaching a green fluorescent protein to the surface of the nano-diamond.
The second, more challenging, specification of the project was that we do not want to have to perfect a new conjugation procedure for every different protein we wish to attach to the diamond as this can be tricky and time consuming. Hence our aim was to optimise the process of attaching a single protein to the diamond. This protein by itself will not be very useful, but will have the property that it will attach incredibly strongly to a second protein in the same way that only the right key will fit in the right lock. Hence the aim was to produce lots of protein covered nano-diamonds, and then generically produce any protein of interest, pre-bound to the other half of the linker molecule. Finally, all we have to do is mix the two products together and we will end up with our required nano-particle covered in an active peptide of choice. This all sounds like a very complicated and long-winded way of doing things, however, it is, in fact, about the easiest way that ensures that all the components will work at the end. The image, right, shows a schematic of the scheme envisaged, here attaching a green fluorescent protein to the surface of the nano-diamond.
 The protein pair at the heart of all this work was developed by our collaborators and is known as Barnase-Barnstar. Each of the two proteins (Barnase and Barnstar, or Bn, Bs for short) are small and robust, being able to exist at high temperature and Ph, and were originally extracted from bacteria, which is useful as it means they do not have any unwanted side effects when being used in mammalian cells. Further to this, when mixed together the 2 proteins form an incredibly strong bond, making them almost inseparable. Both Bn and Bs are again genetically engineered by our collaborators and can be constructed to also contain other active parts such as a dye (Green Fluorescent Protein, GFP, in this case), or again our favourite neuro-peptide, Somatostatin. The aim of the research is thus to attach Barnstar to nano-diamonds and then confirm that Barnase in solution still attaches firmly to the Barnstar when mixed together.
The protein pair at the heart of all this work was developed by our collaborators and is known as Barnase-Barnstar. Each of the two proteins (Barnase and Barnstar, or Bn, Bs for short) are small and robust, being able to exist at high temperature and Ph, and were originally extracted from bacteria, which is useful as it means they do not have any unwanted side effects when being used in mammalian cells. Further to this, when mixed together the 2 proteins form an incredibly strong bond, making them almost inseparable. Both Bn and Bs are again genetically engineered by our collaborators and can be constructed to also contain other active parts such as a dye (Green Fluorescent Protein, GFP, in this case), or again our favourite neuro-peptide, Somatostatin. The aim of the research is thus to attach Barnstar to nano-diamonds and then confirm that Barnase in solution still attaches firmly to the Barnstar when mixed together.
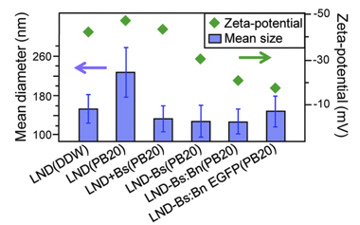
As usual in science we started with more straightforward ideas and build up. One of the biggest problems with nano-particles is that they aggregate, and so rather than having single small particles what is left is a few large clumps. The exact state of the liquid the particles are in can have a large effect on this, for example changing the salt content of the water slightly, or altering the Ph can be the difference between beautiful single particles and a complete mess that is no use to anyone. Hence in these experiments it is very important to keep track of the particles size to make sure the chemical reactions performed have not caused aggregation. The graphs right show the size and charge of nano-diamonds under different situations throughout a chemical reaction. It should be noted that in all cases the size is around 140 nm, the size of single particles except in the case labelled LND(PB20), here the diamonds become unstable by themselves and start to aggregate, however when Barnstar is chemically attached to the surface the size, LND-Bs(PB20) the size drops back, indicating that (a) there has been a reaction as something has changed, and (b) the new particles are stable. By checking the charge of the particles it is also possible to see that something has happened when the reaction is performed. It is very easy to mix things together and hope for the right results, but very hard to full validate and prove that what has happened is what was required, this is made harder at the nano-scale where nothing can be seen by eye. The above plot indicates that the reaction has taken place, but is not proof, and we do not know if the Barnstar is still active or if it has been damaged.
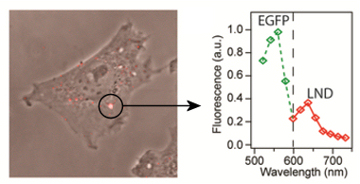 To perform this check, we add our special Bn-GFP to the solution of Bs-Nano-diamonds, and then filter out all the free proteins. Hence what should be left are nano-diamonds, that fluoresce in the red, covered with GFP, that fluoresces in the green, linked together by the Bs-Bn bond. To add a further challenge, we wish to ensure that this new hybrid particle can also survive being consumed by a cell. The final picture, right, shows a cell with a single GFP coated nano-diamond, the spectra confirms this is the case and not just something yellow that looks similar.
To perform this check, we add our special Bn-GFP to the solution of Bs-Nano-diamonds, and then filter out all the free proteins. Hence what should be left are nano-diamonds, that fluoresce in the red, covered with GFP, that fluoresces in the green, linked together by the Bs-Bn bond. To add a further challenge, we wish to ensure that this new hybrid particle can also survive being consumed by a cell. The final picture, right, shows a cell with a single GFP coated nano-diamond, the spectra confirms this is the case and not just something yellow that looks similar.
By performing many other experiments, it is possible to prove that all the aspects of these proteins and reactions have worked and we are left with the situation shown schematically at the top of the page. The final step is to replace the Bn-GFP with Bn-SST, and we have a diamond that should follow the somatostatin receptors inside living cells.
As a further step we have shown that the Bn:Bs system can be used to for bio-assay techniques. Assays are used to detect the presence of trace levels of certain biological agents but can suffer from the inherent level of noise in the system when very low levels of the proteins need to be detected. Bn:Bs can be useful in certain situations when blood samples need to be analysed as the bacterial origin of the proteins mean that they interacts minimally with mammalian proteins. This means the Bn:Bs pair will suffer lower levels of non-specific binding, providing a lower noise floor and hence a potentially higher sensitivity. Early testes indicated that this could be viable paving the way for further work.
 Barstar:barnase - a versatile platform for colloidal diamond bioconjugation
Barstar:barnase - a versatile platform for colloidal diamond bioconjugation
V.K.A. Sreenivasan, E.A. Ivukina, W. Deng, T.A. Kelf, T.A. Zdobnova, S.V. Lukash, B.V. Veryugin, O.A. Stremovskiy, A.V. Zvyagin, and S.M. Deyev
J. Mat. Chem.21, 65 (2011)
 A modular design of low-background bioassays based on a high-affinity molecular pair barstar:barnase
A modular design of low-background bioassays based on a high-affinity molecular pair barstar:barnase
V.K.A. Sreenivasan, T.A. Kelf, E.A. Grebenik, O.A. Stremovskiy, J.M. Say, J.R. Rabeau, A.V. Zvyagin, S.M. Deyev.
Proteomics. 13, 1737-1443 (2013)
In the non-specific endocytosis section an understanding was gained about how to stop nano-particles from entering cells when unwanted, and in the receptor mediated endocytosis section there was a discussion on how a genetically engineered protein called SST was tested and found to travel the same specific cellular pathways as the un-engineered protein, but to also have a second, fluorescent protein attached to it (RFP). As the newly created protein is genetically engineered it is a single protein that is both SST and RFP, and hence will not, for example, fall apart. It was also tested and proven to work efficiently to label SST receptors inside cells. The final test, therefore, was to attach a similar protein to a nano-particle. While this may sound easy, even a slight alteration to the protein structure can destroy the SST's activity. On top of this, we also have some other quite difficult specifications. As a nano-particle we wish to use a nano-diamond. This might sound extravagant but nano-sized pieces of diamond can be produced in large quantities but simply blowing up TNT (making them cheap), and tiny diamonds have many nice features. First, they are pure carbon, so are inert and non-toxic to cells, second, they can emit light through things called colour centres, and these are not damaged by the cells or bleached in time (as is the case for dyes). Finally, the carbon surfaces of the diamonds are a nice starting point for chemical bonding reactions.
 The second, more challenging, specification of the project was that we do not want to have to perfect a new conjugation procedure for every different protein we wish to attach to the diamond as this can be tricky and time consuming. Hence our aim was to optimise the process of attaching a single protein to the diamond. This protein by itself will not be very useful, but will have the property that it will attach incredibly strongly to a second protein in the same way that only the right key will fit in the right lock. Hence the aim was to produce lots of protein covered nano-diamonds, and then generically produce any protein of interest, pre-bound to the other half of the linker molecule. Finally, all we have to do is mix the two products together and we will end up with our required nano-particle covered in an active peptide of choice. This all sounds like a very complicated and long-winded way of doing things, however, it is, in fact, about the easiest way that ensures that all the components will work at the end. The image, right, shows a schematic of the scheme envisaged, here attaching a green fluorescent protein to the surface of the nano-diamond.
The second, more challenging, specification of the project was that we do not want to have to perfect a new conjugation procedure for every different protein we wish to attach to the diamond as this can be tricky and time consuming. Hence our aim was to optimise the process of attaching a single protein to the diamond. This protein by itself will not be very useful, but will have the property that it will attach incredibly strongly to a second protein in the same way that only the right key will fit in the right lock. Hence the aim was to produce lots of protein covered nano-diamonds, and then generically produce any protein of interest, pre-bound to the other half of the linker molecule. Finally, all we have to do is mix the two products together and we will end up with our required nano-particle covered in an active peptide of choice. This all sounds like a very complicated and long-winded way of doing things, however, it is, in fact, about the easiest way that ensures that all the components will work at the end. The image, right, shows a schematic of the scheme envisaged, here attaching a green fluorescent protein to the surface of the nano-diamond. The protein pair at the heart of all this work was developed by our collaborators and is known as Barnase-Barnstar. Each of the two proteins (Barnase and Barnstar, or Bn, Bs for short) are small and robust, being able to exist at high temperature and Ph, and were originally extracted from bacteria, which is useful as it means they do not have any unwanted side effects when being used in mammalian cells. Further to this, when mixed together the 2 proteins form an incredibly strong bond, making them almost inseparable. Both Bn and Bs are again genetically engineered by our collaborators and can be constructed to also contain other active parts such as a dye (Green Fluorescent Protein, GFP, in this case), or again our favourite neuro-peptide, Somatostatin. The aim of the research is thus to attach Barnstar to nano-diamonds and then confirm that Barnase in solution still attaches firmly to the Barnstar when mixed together.
The protein pair at the heart of all this work was developed by our collaborators and is known as Barnase-Barnstar. Each of the two proteins (Barnase and Barnstar, or Bn, Bs for short) are small and robust, being able to exist at high temperature and Ph, and were originally extracted from bacteria, which is useful as it means they do not have any unwanted side effects when being used in mammalian cells. Further to this, when mixed together the 2 proteins form an incredibly strong bond, making them almost inseparable. Both Bn and Bs are again genetically engineered by our collaborators and can be constructed to also contain other active parts such as a dye (Green Fluorescent Protein, GFP, in this case), or again our favourite neuro-peptide, Somatostatin. The aim of the research is thus to attach Barnstar to nano-diamonds and then confirm that Barnase in solution still attaches firmly to the Barnstar when mixed together.
As usual in science we started with more straightforward ideas and build up. One of the biggest problems with nano-particles is that they aggregate, and so rather than having single small particles what is left is a few large clumps. The exact state of the liquid the particles are in can have a large effect on this, for example changing the salt content of the water slightly, or altering the Ph can be the difference between beautiful single particles and a complete mess that is no use to anyone. Hence in these experiments it is very important to keep track of the particles size to make sure the chemical reactions performed have not caused aggregation. The graphs right show the size and charge of nano-diamonds under different situations throughout a chemical reaction. It should be noted that in all cases the size is around 140 nm, the size of single particles except in the case labelled LND(PB20), here the diamonds become unstable by themselves and start to aggregate, however when Barnstar is chemically attached to the surface the size, LND-Bs(PB20) the size drops back, indicating that (a) there has been a reaction as something has changed, and (b) the new particles are stable. By checking the charge of the particles it is also possible to see that something has happened when the reaction is performed. It is very easy to mix things together and hope for the right results, but very hard to full validate and prove that what has happened is what was required, this is made harder at the nano-scale where nothing can be seen by eye. The above plot indicates that the reaction has taken place, but is not proof, and we do not know if the Barnstar is still active or if it has been damaged.
 To perform this check, we add our special Bn-GFP to the solution of Bs-Nano-diamonds, and then filter out all the free proteins. Hence what should be left are nano-diamonds, that fluoresce in the red, covered with GFP, that fluoresces in the green, linked together by the Bs-Bn bond. To add a further challenge, we wish to ensure that this new hybrid particle can also survive being consumed by a cell. The final picture, right, shows a cell with a single GFP coated nano-diamond, the spectra confirms this is the case and not just something yellow that looks similar.
To perform this check, we add our special Bn-GFP to the solution of Bs-Nano-diamonds, and then filter out all the free proteins. Hence what should be left are nano-diamonds, that fluoresce in the red, covered with GFP, that fluoresces in the green, linked together by the Bs-Bn bond. To add a further challenge, we wish to ensure that this new hybrid particle can also survive being consumed by a cell. The final picture, right, shows a cell with a single GFP coated nano-diamond, the spectra confirms this is the case and not just something yellow that looks similar.
By performing many other experiments, it is possible to prove that all the aspects of these proteins and reactions have worked and we are left with the situation shown schematically at the top of the page. The final step is to replace the Bn-GFP with Bn-SST, and we have a diamond that should follow the somatostatin receptors inside living cells.
As a further step we have shown that the Bn:Bs system can be used to for bio-assay techniques. Assays are used to detect the presence of trace levels of certain biological agents but can suffer from the inherent level of noise in the system when very low levels of the proteins need to be detected. Bn:Bs can be useful in certain situations when blood samples need to be analysed as the bacterial origin of the proteins mean that they interacts minimally with mammalian proteins. This means the Bn:Bs pair will suffer lower levels of non-specific binding, providing a lower noise floor and hence a potentially higher sensitivity. Early testes indicated that this could be viable paving the way for further work.
Papers relating to this work:
 Barstar:barnase - a versatile platform for colloidal diamond bioconjugation
Barstar:barnase - a versatile platform for colloidal diamond bioconjugation
V.K.A. Sreenivasan, E.A. Ivukina, W. Deng, T.A. Kelf, T.A. Zdobnova, S.V. Lukash, B.V. Veryugin, O.A. Stremovskiy, A.V. Zvyagin, and S.M. Deyev
J. Mat. Chem.21, 65 (2011)
 A modular design of low-background bioassays based on a high-affinity molecular pair barstar:barnase
A modular design of low-background bioassays based on a high-affinity molecular pair barstar:barnase
V.K.A. Sreenivasan, T.A. Kelf, E.A. Grebenik, O.A. Stremovskiy, J.M. Say, J.R. Rabeau, A.V. Zvyagin, S.M. Deyev.
Proteomics. 13, 1737-1443 (2013)